Comprehensive analysis of the aging dynamics across the mouse lifespan.
About
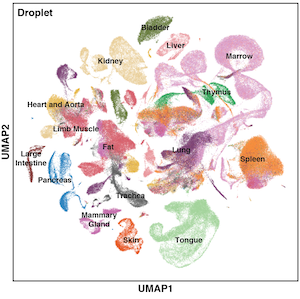
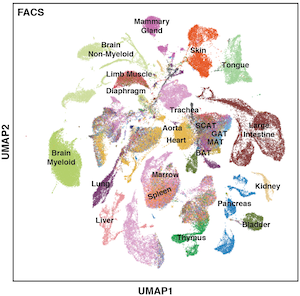
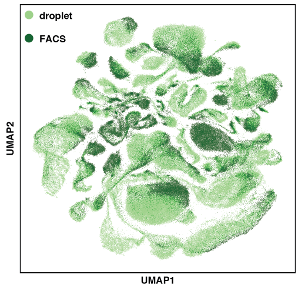
The Tabula Muris Senis is a comprehensive resource for the cell biology community which offers a detailed molecular and cell-type specific portrait of aging. We view such cell atlas as an essential companion to the genome: the genome provides a blueprint for the organism but does not explain how genes are used in a cell type specific manner or how the usage of genes changes over the lifetime of the organism. The cell atlas provides a deep characterization of phenotype and physiology and serves as a reference for understanding many aspects of the cell biological changes that mammals undergo during their lifespan. The project includes single-cell and bulk RNA-sequencing of different organs across the mouse lifespan.
Publications
The Tabula Muris Consortium, Nature (2020)
The Tabula Muris Consortium, Nature (2022)
Pálovics et al., Nature (2022)
Nicholas Schaum et al., Nature (2020)
Zhang et al., eLife (2021)
Complete Dataset
Datasets split by assay
Tissue collection available in cellxgene

Raw Data Available on AWS

The raw sequencing files of the Tabula Muris Senis projects are made available for anyone to use via Amazon S3

Annotated and/or partially annotated objects to use with scanpy and cellxgene are available from figshare